Re-Engineering CI/CD pipelines for SciPy
In this blog post I talk about the projects and my work during my internship at Quansight Labs. My efforts were geared towards re-engineering CI/CD pipelines for SciPy to make them more efficient to use with GitHub Actions. I also talk about the milestones that I achieved, along with the associated learnings and improvements that I made.
This blog post would assume a basic understanding of CI/CD and GitHub Actions. I will also assume a basic understanding of Python and the SciPy ecosystem.
 Re-Engineering CI/CD pipelines for SciPy
Re-Engineering CI/CD pipelines for SciPy
SciPy is quite an old and mature project, which is being used across the PyData community. The project currently uses a variety of continuous integration services and the Setuptools build system. As the project is growing and distutils will be deprecated as part of PEP 632, it was reasonable to migrate to a new build system. A previous blog post describes the migration process in detail and sets the future course for SciPy developers to look forward to faster builds.
SciPy has been previously using TravisCI, CircleCI, and AppVeyor including GitHub Actions across its continuous integration pipelines.My project was centered around building and re-engineering the continuous integration pipelines for SciPy using GitHub Actions to be made compatible with the new build system and increasing the adoption of GitHub Actions.
Why GitHub Actions?
GitHub Actions features a vast number of architectures, operating systems, runners, and more. With the aid of GitHub Actions, we can directly use the GitHub API from our workflows themselves. GitHub Actions also features a vibrant marketplace with applications centered around making the usage of GitHub Actions more straightforward for developers.
We can also emulate local CI runs using a tool like Act to validate our GitHub Actions workflow. It allows us to run the entire workflow (including build and test) on our local machine, using a Docker image without initially testing it on the CI. Apart from this, whenever we fork a repository, the actions automatically get forked. It allows us to test on our fork and makes it relatively straightforward to use.
 Life is good but it can be better with GitHub Actions
Life is good but it can be better with GitHub Actions
Project Plan
I sketched the project plan through the inputs provided by Ralf Gommers and my mentor John Lee. To document the entire project plan, we had to spend a couple of weeks researching and brainstorming on the existing CI pipelines for SciPy and what can be the new additions around the same. The list below describes them.
-
SciPy uses markdown templates for reporting bugs and feature requests through Issues. We can migrate them to use GitHub Issue forms in YAML for a better user experience.
-
SciPy CI build time takes around ~10 minutes with the Meson build system. It is an improvement over the existing
setuptools,but for faster iterations, we need to capitalize on caching to reduce the total run-time of CI. -
SciPy is currently tested on a variety of operating systems and architecture. We want to ensure the compatibility of the new build system for the same. A CI matrix should define the combination of variations on the build and the ideal target.
-
SciPy currently features benchmarks that are not automated on the CI. We want to automate this process on the CI itself by tagging Pull Requests with specific labels or triggering the same during releases.
-
Scipy currently supports x86, x86_64, PPC64, and ARM64 architecture through the Azure DevOps pipelines. We want to ensure that the compatibility remains the same with the new build system.
-
SciPy uses parallel builds across the Azure DevOps pipelines and macOS GitHub Actions along with the test scripts. We want to consider what parts of the entire matrix we wish to run on the CI providers we choose to balance the overhead of using multiple providers.
-
SciPy currently lacks any developer documentation for CI providers. Documenting the CI systems and the build matrix is necessary for developers to cross-reference them in future improvements. We can also document how to emulate CI runs locally through specific tools.
Implementing the local CI run
The project's first step was to ensure that we could run the GitHub Actions CI locally to ensure faster iterations on the failures we encountered. GitHub does not feature an official tool to make this possible; hence we had to look at third-party alternatives like Act.
act is a tool offered by Nektos which provides a handy way to run GitHub Actions locally. It gives a quick way to validate your changes on the CI locally, without committing/pushing your changes to the workflows to trigger and validate them. It leads to fast feedback and compatibility as a local task runner to validate all our CI jobs.
You can try out act yourself by cloning Ralf’s fork and referencing the documentation for the act usage. It keeps the GitHub Action notifications clutter-free and reduces the chance of accidentally triggering any failure during debugging.
Here is act in action with the meson branch over Ralf’s fork:
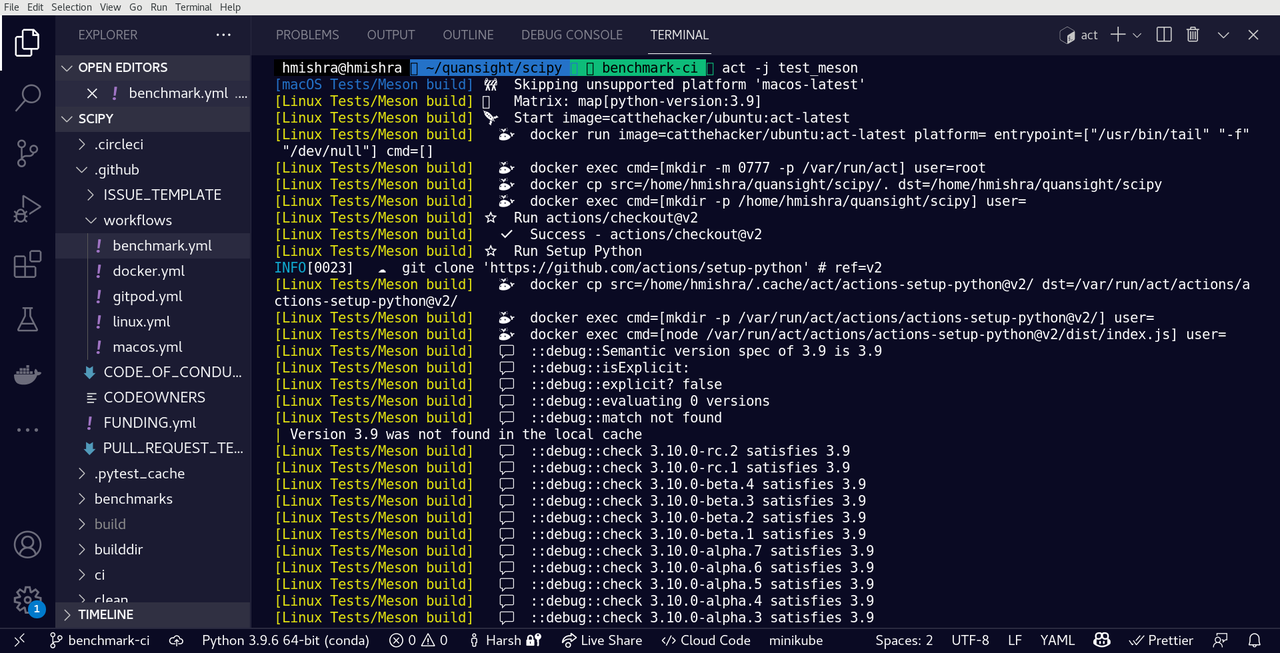 Usage of `act` with GitHub Actions for SciPy
Usage of `act` with GitHub Actions for SciPy
To ensure that we can test the GitHub Actions locally, we had to make a lot of tweaks to ensure that act plays nicely with local workflows. PR #57 containing the commits for the tweaks and the overall developer documentation for the act usage.
Generating dynamic files in the build
During the Meson build process, we generate some of the dynamic files during the process. The overall plan was to generate these files programmatically and incorporate this into the build in the most sensible way. The files that we were looking to generate dynamically were:
-
__config__.py: This file is generated by NumPy and should use a version of NumPy at build time that is compatible with all potential runtime versions of NumPy. -
version.py: This file is dependent on the Git commit hash. We needed to generate a file in the build directory instead of in-place, because Meson does not allow writing output into the source tree that is being built.
This task was specifically a challenging one, given my lack of experience with Meson build system. After understanding some of the specifics around it and digging deep into the documentation, I tackled it. We can use Meson’s custom_target to dynamically generate these files during the build time.
With the help of this, we were able to remove the hard-coded __config__.py and version.py from the source directory and ensure a clean build and install. PR #57 contains the commits for the code required to generate the dynamic files and associated refactors in the setup.py file.
Integrating build caching using Ccache
Today, it takes around ~35 minutes for SciPy's Linux and macOS tests to pass on GitHub Actions. With Scipy’s migration efforts towards the Meson build system, it takes around ~25 minutes for the CI to pass the build, install, and run tests. Our primary focus was to reduce the overall time to achieve this. But finding workarounds for the same was particularly challenging.
John initially proposed the idea of caching the source and the build directory together. We can restore the cache during the workflow and use remote sync to update the subset of changed files. It would allow us to perform an incremental build.
While discussing with Ralf, I realized that using timestamp-based caching would be the way to go forward. My initial approach was to cache the build directory, which was nowhere effective. My conversation with Meson developers helped me realize that using a compiler caching tool like ccache or sccache would be the way forward to cache the build targets.
To make sure that dependencies are being cached, I capitalized on the cache action. GitHub Workflow runs often reuse the same downloaded dependencies from one run to another. GitHub’s cache actions allow us to retrieve cache identified by a unique key, for which I used a unique timestamp-based caching approach.
While digging around the documentation, I discovered that ccache is natively supported by Meson and hence would be a better choice. I found that the build targets are exactly being cached by debugging around, and I can use the cache action to cache them. Finally, I made it work, thus effectively reducing the build time by nearly ~70% and reducing the CI time.
In addition, I also worked on caching PyPI dependencies using the cache actions that allowed us to shave off some of the time required during the Python dependency installation. PR #63 containing the commits for the code necessary to set up build caching as part of the GitHub Action workflow for Linux.
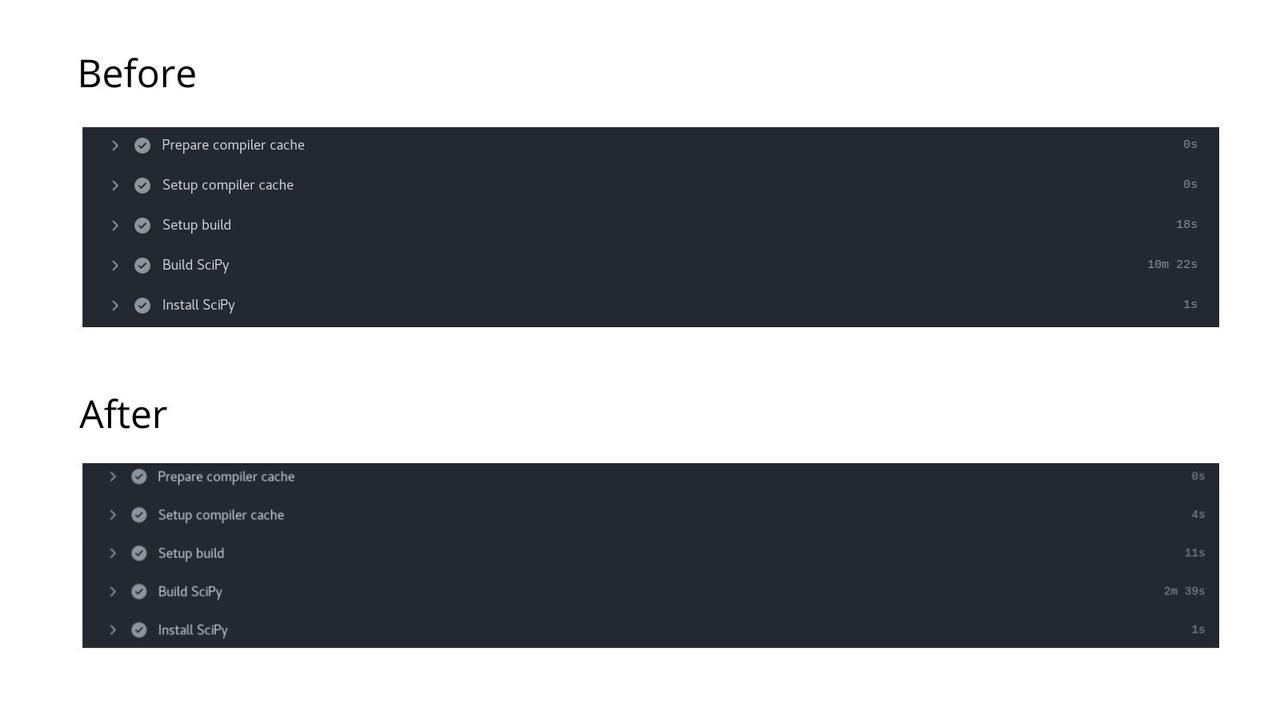 Implementation of Ccache with SciPy's Meson build system to reduce CI build time.
Implementation of Ccache with SciPy's Meson build system to reduce CI build time.
Implementing CI job for macOS
With the GitHub Action workflows available for Linux, it seemed reasonable that we would like to test the same on macOS. This issue by Rohit Goswami, where he documents the problems with the macOS build, made sense that we can implement a workflow with little to almost no tweaks. However, I wanted to implement the conda-based approach rather than putting forward a native build.
GitHub Actions allows us to perform macOS builds quite generously, and creating a workflow for the same was relatively easy. After this, we had to make a couple of iterations to verify if the compiler caching exactly works and how performant it is compared to a fresh build. I also implemented conda caching for faster installs using Mamba, which reduced the effective build time by 30%.
PR #65 containing the commits for the code required to set up the GitHub Action workflow for macOS with compiler caching and conda caching.
Automating CI benchmarking on GitHub Actions
This task was heavily influenced by the work done by Jaime Rodríguez-Guerra on Scikit-Image PR #5424. We wanted the benchmarks to be run automatically on specific Pull Requests with apt labels or during releases. It, however had a few problems that we wanted to address before moving forward:
-
Benchmarking requires to happen on specific hardware under specific conditions, which is difficult to reproduce.
-
Benchmarking on CI is even harder, primarily when different runners are provided with shared resources.
-
Having dedicated hardware for the same can lead to abuse and unnecessary noise.
We decided to use either Cachegrind or Jaime’s approach to tackle all of these problems, which talks about relative benchmarking on GitHub Actions. Both scikit-image and SciPy use Airspeed Velocity (asv) for benchmarking. This rules out the use of cachegrind since asv is not compatible with the same.
Following in Jaime’s footsteps, I used asv’s subcommand called continuous to run a benchmark test on two corresponding commits. This will collect all the statistics together and give us a log and results, which can be uploaded as an artifact after the workflow gets over. It would allow the maintainers to directly download the same and verify on their local machine before moving ahead with a pull request or a release.
PR #67 contains the commits for the code required to set up the GitHub Action workflow for automated CI benchmarking using asv and artifact upload.
Strategizing the CI Matrix for SciPy
One of the project milestones was to strategize the CI matrix for SciPy, which can be distributed across GitHub Actions, Azure DevOps, and CircleCI. To account for the same, we spent significant time identifying the existing CI for SciPy and finding all the operating systems, architecture, and special purpose workflows that we wish to support.
After a lot of iterations and discussions with Ralf, we were able to identify the following operating systems/architecture to be supported:
-
Linux (32-bit X86 and 64-bit X86): Supported across GitHub Actions Nightly build, pull requests, and Azure DevOps.
-
Linux (aarch64): Supported on a GitHub Action workflow for pull requests.
-
Linux (PPC64): Supported on a GitHub Action workflow for pull requests.
-
Windows (32-bit and 64-bit): Requires further efforts for the Meson build using MSVC and Gfortran required for building wheels.
-
macOS (X86): Supported on a GitHub Action workflow for pull requests.
-
macOS (ARM64): Requires more effort with no options available on GitHub Actions.
We also implemented a sampling strategy to match the versions of Python and NumPy. In addition, the special purpose jobs included are for linting, docs build, Gitpod build, benchmarking, Pythran, MyPy, and more.
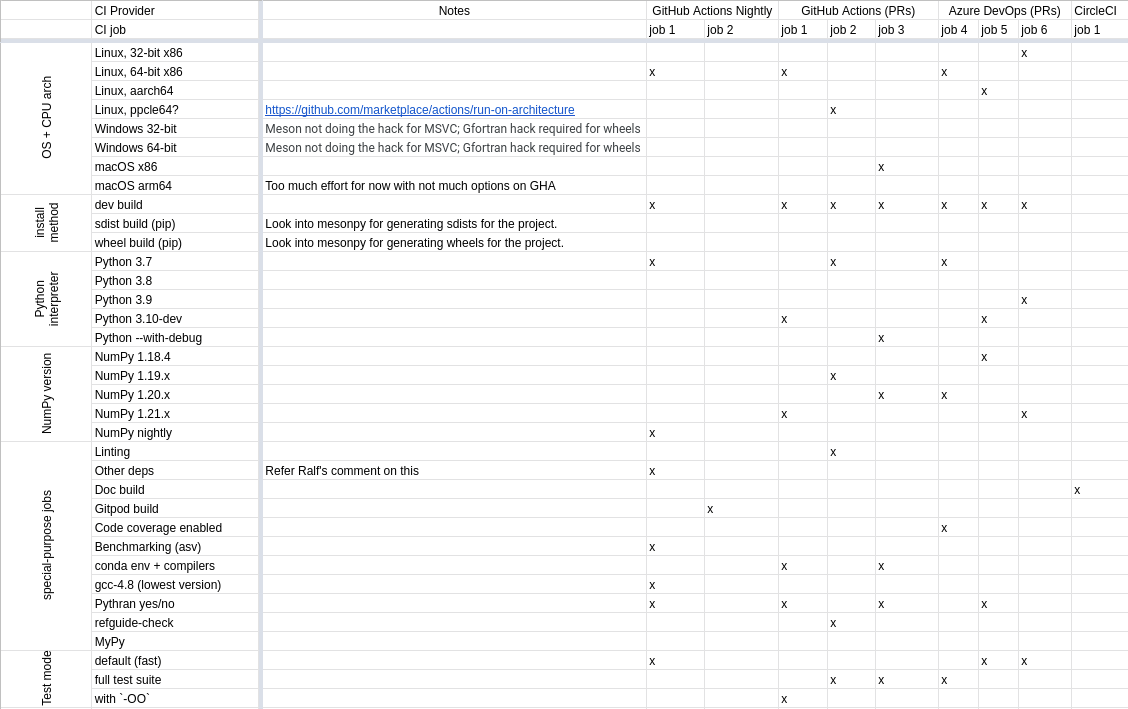 Spreadsheet displaying the CI matrix for the SciPy Meson build which utilizes GitHub Actions, Azure DevOps, and CircleCI.
Spreadsheet displaying the CI matrix for the SciPy Meson build which utilizes GitHub Actions, Azure DevOps, and CircleCI.
Miscellaneous
In addition, I also worked on the upstream SciPy project on some of the minor issues. PR #14993 updates all the Issue markdown templates to utilize the new YAML-based GitHub issue forms. The feedback received after the previous pull request was incorporated on PR #14669.
Challenges faced
While this whole project was challenging, some aspects of it were unforeseen and more challenging. When I decided to go on with this project, I had enough GitHub Actions knowledge to write the pipelines for small projects. I had not enough experience to work with complex GitHub Actions workflows on this big scale. With support from mentors, I was able to overcome this challenge with flying colors.
The other challenge was understanding the Meson build system and how it is being used to build a complex project like SciPy with compiled dependencies. My experience with build systems was limited to setuptools and NPM for Python and JavaScript, respectively. After digging through a lot of extensive documentation, I figured out the nits and grits around the Meson build system.
The most challenging task was to make the workflows work nicely with GitHub Actions. It was associated with many debugging sessions using Tmate, which took me the most time. All thanks to my mentor John who helped me hack through the CI workflows and the build system in no time.
Future Plans
There is a lot to do with the new CI matrix and the configurations, and it will take more time to reach the desired maturity. I was able to meet most of the goals, but some of them remain.
-
Implement the CI matrix on the GitHub Actions workflow.
-
Add associated developer documentation for CI matrix and service providers.
-
Improve and optimize the project as it is tested on multiple architectures.
-
Automate source and binary distribution through GitHub releases.
-
Add GitHub Actions for build and uploading SciPy wheels through a custom GitHub repository.
What did I learn from this project?
The summer internship at Quansight Labs has been the busiest time of my life for all good reasons. I learned a great deal about GitHub Actions, build systems, and PyData projects through the same. I also learned about testing and benchmarking, along with a more in-depth understanding of Docker. I had the basic idea for some of them and have worked on them in hobby projects, but implementing them myself on a significant project and fixing bugs was a wholesome experience.
Apart from the same, I learned a great deal about the communication side of things. Communicating with my mentor and the stakeholders improved my thought process and intuition behind how things work. It also helped me instill a “doer” mentality, where I work fast, fail fast, and iterate quickly. Getting to know fellow Interns and the great work they have been doing was also significant for me to understand the ecosystem.
Acknowledgements
My internship at Quansight Labs has been one of the most wholesome experiences I ever had. I’m grateful to have worked on this project and get acquainted with Open Source, being in my undergraduate studies itself. Open Source has taught me a great deal about many things, and this project has further paved the way for me to cherish my ambitions of working more on the Python and DevOps side of things.
I want to thank my mentor John, for his impeccable mentorship and feedback. Without his help and support, all this would not have been possible. Above all, his skill in drawing from past experiences to future model outcomes has been helpful for me to understand things beyond open source and software development itself.
I would also like to thank Ralf, who has been instrumental in my project work. His feedback, continued support, and long-term vision contributed significantly to my project. Anirudh, my co-intern, helped me understand more about SciPy and helping with my first PR reviews. Jaime helped me considerably understand benchmarking, and getting to know more about his work inspired me. They all are very polite, knowledgeable, and helpful.
Thanks to the SciPy community and Quansight Labs for this fantastic experience!
References
This section contains references to the articles, blogs, and websites that I used to learn more about the project:
Comments